3D Pathology
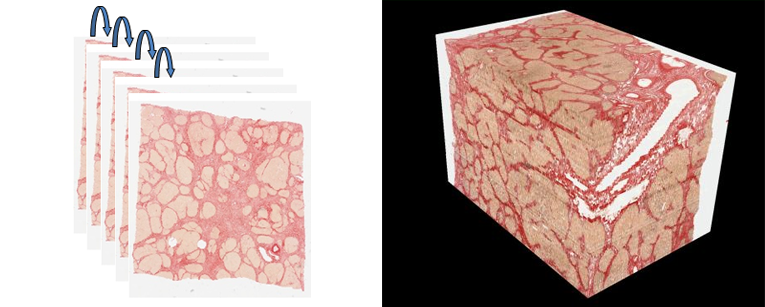
The HG-MIM 3D Pathology Add-on enables the stacking up of hundreds of serial section images to form a volumetric dataset. The add-on supports:
-
Rigid and Non-rigid alignment (for correction of sectioning induced warping)
-
Annotation of volumetric data at full resolution using the HG-MIM pan and zoom interface (curves, ellipses, boxes, lines, arrows, etc.).
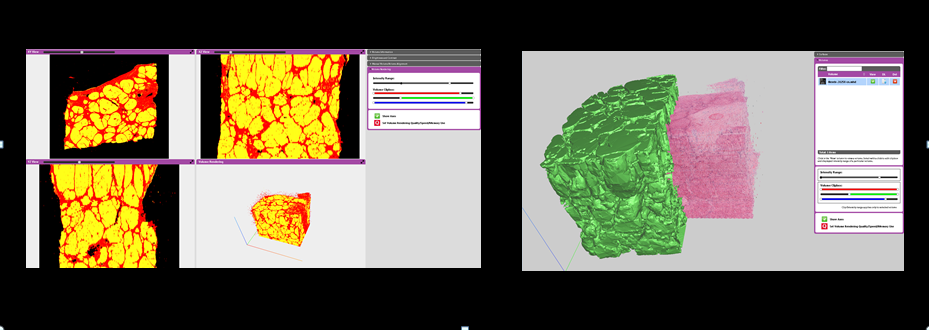
Annotations of a image stack can be converted into 3D surface models, visualised within a web browser, and exported in STL format (or exported as binary volumes in MHD format).
-
Generation of volumetric data in standard volumetric formats (MHD, Analyze etc.) for visualisation HG-MIM Volume Viewer, or other standard volume viewing tools.
-
Conversion of annotations into volumetric data for 3D rendering.
-
Conversion of annotations from/to original image frame of reference from/to 3D volumetric frame of reference (including imported annotations e.g from Aperio ImageScope).
-
Colour correction (to compensate for stain variation between slides)
- Integration with HG-MIM Deep Learning add-on allowing classified volumetric data sets and surfaces to be generated (see below).
If you are interested in alignment of pathology volumes to radiology data, or blockface imaged volumes why not take a look at our Pathology:Radiology Add-on. This supports multiple slices and is fully compatible with the 3D pathology add-on.
Additionally, our Multi-stain Add-on also supports image stacks, and is fully compatible with the 3D pathology add-on.

